
Finding suggests genomes may be rife with hidden similarities
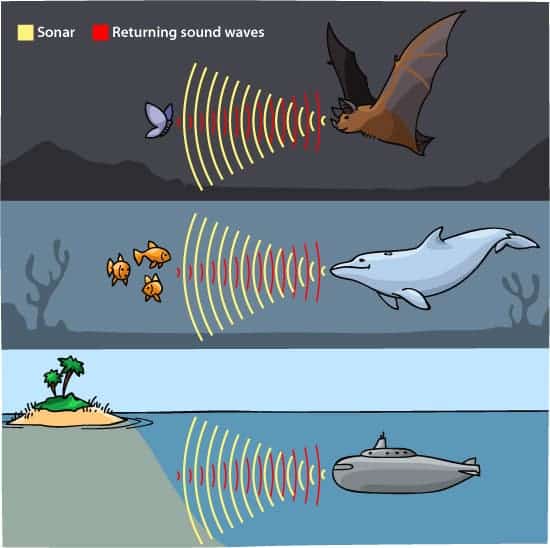
Dolphins and bats don’t have much in common, but they share a superpower: Both hunt their prey by emitting high-pitched sounds and listening for the echoes. Now, a study shows that this ability arose independently in each group of mammals from the same genetic mutations. The work suggests that evolution sometimes arrives at new traits through the same sequence of steps, even in very different animals. The research also implies that this convergent evolution is common—and hidden—within genomes, potentially complicating the task of deciphering some evolutionary relationships between organisms.

Nature is full of examples of convergent evolution, wherein very distantly related organisms wind up looking alike or having similar skills and traits: Birds, bats, and insects all have wings, for example. Biologists have assumed that these novelties were devised, on a genetic level, in fundamentally different ways. That was also the case for two kinds of bats and toothed whales, a group that includes dolphins and certain whales, that have converged on a specialized hunting strategy called echolocation. Until recently, biologists had thought that different genes drove each instance of echolocation and that the relevant proteins could change in innumerable ways to take on new functions.

But in 2010, Stephen Rossiter, an evolutionary biologist at Queen Mary, University of London, and his colleagues determined that both types of echolocating bats, as well as dolphins, had the same mutations in a particular protein called prestin, which affects the sensitivity of hearing. Looking at other genes known to be involved in hearing, they and other researchers found several others whose proteins were similarly changed in these mammals.

Now, Rossiter’s team has expanded the search for this so-called molecular convergence to the entire genome. They sequenced the genomes of four species from various branches of the bat family tree, two that use echolocation and two that don’t. They added in the existing genome sequences of the large flying fox and the little brown bat, another echolocator. Evolutionary biologist Joe Parker, also at Queen Mary, University of London, compared the bat genetic sequences to those from more than a dozen other mammals, including the bottlenose dolphin. He focused on the 2300 genes that exist in single copies in all the bats, the dolphin, and at least five other mammals. He evaluated how similar each gene was to its counterparts in various bats and the dolphin. The analysis revealed that 200 genes had independently changed in the same ways, Parker, Rossiter and their colleagues report today in Nature. Several of the genes are involved in hearing, but the others have no clear link to echolocation so far; some genes with shared changes are important for vision, but most have functions that are unknown.
“The biggest surprise,” says Frédéric Delsuc, a molecular phylogeneticist at Montpellier University in France, “is probably the extent to which convergent molecular evolution seems to be widespread in the genome.”
Genomicist Todd Castoe from the University of Texas, Arlington, is also impressed: “I’m pretty convinced they are finding something real, and it’s really exciting [and] pretty important.” However, he is critical about the way the analysis was done, suggesting that the approach found only indirect evidence of molecular convergence.

The discovery that molecular convergence can be widespread in a genome is “bittersweet,” Castoe adds. Biologists building family trees are likely being misled into suggesting that some organisms are closely related because genes and proteins are similar due to convergence, and not because the organisms had a recent common ancestor. No family trees are entirely safe from these misleading effects, Castoe says. “And we currently have no way to deal with this.”

Leave a Reply